1.14.19.35: sn-2 acyl-lipid omega-3 desaturase (ferredoxin)
This is an abbreviated version!
For detailed information about sn-2 acyl-lipid omega-3 desaturase (ferredoxin), go to the full flat file.
Word Map on EC 1.14.19.35 
Reaction
 a (7Z,10Z)-hexadeca-7,10-dienoyl-[glycerolipid] +
2 reduced ferredoxin
+
a (7Z,10Z)-hexadeca-7,10-dienoyl-[glycerolipid] +
2 reduced ferredoxin
+
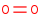 O2 +
2 H+
=
O2 +
2 H+
=
 a (7Z,10Z,13Z)-hexadeca-7,10,13-trienoyl-[glycerolipid] +
2 oxidized ferredoxin
+
2 H2O
a (7Z,10Z,13Z)-hexadeca-7,10,13-trienoyl-[glycerolipid] +
2 oxidized ferredoxin
+
2 H2O
Synonyms
16:2/18:2 desaturase, acyl-lipid omega-3 desaturase, FAD7, FAD7 omega 3-desaturase, FAD7-1, FAD7-2, FAD8, fatty acid desaturase 7, fatty acid desaturase-7, fatty acid desaturase7, GmFAD7, HaFAD7, HvFAD7, omega-3 desaturase, omega-3 FA desaturase, omega-3 fatty acid desaturase, omega-3 fatty acid desaturase gene, omega3 fatty acid desaturase, omega3-fatty acid desaturase, OsFAD8, PfrFAD7-1, PfrFAD7-2, plastid omega-3 desaturase, plastidial FAD7 omega-3 desaturase, plastidial omega-3 desaturase, plastidial omega-3 fatty acid desaturase, Sefad7, sn-2 acyl-lipid omega-3 desaturase (ferredoxin)
ECTree
Sequence
Sequence on EC 1.14.19.35 - sn-2 acyl-lipid omega-3 desaturase (ferredoxin)
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
FAD3C_ARATH
446
3
51174
Swiss-Prot
Chloroplast (Reliability: 2)
FAD3C_HELAN
443
1
50808
Swiss-Prot
Chloroplast (Reliability: 2)
FAD3D_ARATH
435
3
50136
Swiss-Prot
Chloroplast (Reliability: 2)
A0A5B6Z354_DAVIN
334
3
38593
TrEMBL
other Location (Reliability: 5)
A0A5B6Z0F9_DAVIN
236
1
26807
TrEMBL
other Location (Reliability: 5)
A0A812CI62_SEPPH
380
6
44920
TrEMBL
Secretory Pathway (Reliability: 5)
A0A192GPD1_9MAGN
450
3
51589
TrEMBL
Chloroplast (Reliability: 2)
A0A7R8H7D8_LEPSM
237
3
26855
TrEMBL
other Location (Reliability: 2)
A0A5B6Z1X1_DAVIN
432
3
49702
TrEMBL
other Location (Reliability: 5)
A0A5B6Z0F2_DAVIN
427
3
49037
TrEMBL
other Location (Reliability: 5)
A0A5B6Z016_DAVIN
427
3
48885
TrEMBL
other Location (Reliability: 5)
B2ZFX8_9CHLO
436
3
49067
TrEMBL
Chloroplast (Reliability: 5)
W0S9D5_FISSO
422
6
48251
TrEMBL
Mitochondrion (Reliability: 5)
W0SDG6_FISSO
421
6
48197
TrEMBL
Secretory Pathway (Reliability: 5)
A0A7R8CBH7_LEPSM
462
5
52915
TrEMBL
other Location (Reliability: 3)
A0A190ZLB9_PERFR
438
0
50028
TrEMBL
-
A8DTI8_POROL
445
0
50449
TrEMBL
-
A8HMC4_CHLRE
418
0
47635
TrEMBL
-
C4MH52_SOYBN
453
0
51247
TrEMBL
-
C6ZGG8_POROL
436
0
50600
TrEMBL
-
L0GGK2_CAMSI
438
0
50136
TrEMBL
-
O23824_MAIZE
443
0
49438
TrEMBL
-
O24626_MAIZE
398
0
44789
TrEMBL
-
FAD3C_SESIN
447
0
51116
Swiss-Prot
-
Q7X7I9_SOLLC
435
0
49660
TrEMBL
-
Q84NP3_ORYSJ
413
0
47012
TrEMBL
-
S5GIT3_CAMSI
452
0
51568
TrEMBL
-
html completed




 results (
results ( results (
results ( top
top









