2.1.1.202: multisite-specific tRNA:(cytosine-C5)-methyltransferase
This is an abbreviated version!
For detailed information about multisite-specific tRNA:(cytosine-C5)-methyltransferase, go to the full flat file.
Word Map on EC 2.1.1.202 
Reaction
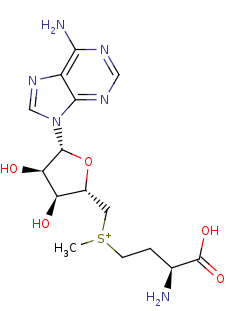 S-adenosyl-L-methionine +
S-adenosyl-L-methionine +
 cytosine34 in tRNA precursor=
cytosine34 in tRNA precursor=
 S-adenosyl-L-homocysteine +
S-adenosyl-L-homocysteine +
 5-methylcytosine34 in tRNA precursor
5-methylcytosine34 in tRNA precursor
Synonyms
5-methylcytosine RNA methyltransferase, broad-spectrum methyltransferase, cytosine-5 RNA methyltransferase, EC 2.1.1.29, HNSun6, M5C RNA methyltransferase, MJ0026, multisite-specific tRNA:m5C-methyltransferase, Ncl1p, NOP2/Sun RNA methyltransferase 2, NSUN2, NSUN6, PhNSUN6, RNA (cytosine-5)-methyltransferase, RNA:m5C methyltransferase, RNA:m5C MTase, TRDMT1, TRM4, TRM4A, TRM4B, Trm4p, tRNA (cytosine72-C5)-methyltransferase, tRNA-specific methyltransferase 4B, tRNA:m5C-methyltransferase, tRNA:m5C72 MTase
ECTree
Sequence
Sequence on EC 2.1.1.202 - multisite-specific tRNA:(cytosine-C5)-methyltransferase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
NCL1_YEAST
Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
684
0
77879
Swiss-Prot
other Location (Reliability: 2)
A0A5B7BVM7_DAVIN
849
0
95253
TrEMBL
Mitochondrion (Reliability: 5)
A0A075GXB8_9EURY
560
0
62351
TrEMBL
-
A0A075HRB1_9EURY
565
0
61467
TrEMBL
-
A0A8J5BK79_9ASCO
745
0
84150
TrEMBL
other Location (Reliability: 1)
A0A0S1XFT8_THEBA
310
0
35427
TrEMBL
-
A0A8J5CLK3_9ASCO
754
0
85020
TrEMBL
other Location (Reliability: 1)
A0A075FID0_9EURY
537
0
58120
TrEMBL
-
A0A2I0BDJ4_9ASPA
921
0
103746
TrEMBL
Mitochondrion (Reliability: 4)
W1QCG0_OGAPD
Ogataea parapolymorpha (strain ATCC 26012 / BCRC 20466 / JCM 22074 / NRRL Y-7560 / DL-1)
639
0
73410
TrEMBL
other Location (Reliability: 2)
A0A0F8B035_CERFI
863
0
96725
TrEMBL
other Location (Reliability: 3)
K0KIE8_WICCF
Wickerhamomyces ciferrii (strain ATCC 14091 / BCRC 22168 / CBS 111 / JCM 3599 / NBRC 0793 / NRRL Y-1031 F-60-10)
691
0
79656
TrEMBL
other Location (Reliability: 2)
A0A396GN75_MEDTR
91
0
10525
TrEMBL
Secretory Pathway (Reliability: 2)
A0A1G4HY69_TRYEQ
806
0
88697
TrEMBL
Mitochondrion (Reliability: 3)
A0A3S7WV22_LEIDO
848
0
92110
TrEMBL
other Location (Reliability: 5)
TRM4B_SCHPO
Schizosaccharomyces pombe (strain 972 / ATCC 24843)
685
0
77822
Swiss-Prot
-
TRDMT_HUMAN
391
0
44597
Swiss-Prot
-
TRDMT_MOUSE
415
0
46794
Swiss-Prot
other Location (Reliability: 4)
NSUN6_PYRHO
Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3)
389
0
44284
Swiss-Prot
-
NSUN2_HUMAN
767
0
86471
Swiss-Prot
other Location (Reliability: 1)
NSUN6_HUMAN
469
0
51770
Swiss-Prot
-
TRM4A_SCHPO
Schizosaccharomyces pombe (strain 972 / ATCC 24843)
688
0
78515
Swiss-Prot
-
html completed




 results (
results ( results (
results ( top
top









