2.4.1.50: procollagen galactosyltransferase
This is an abbreviated version!
For detailed information about procollagen galactosyltransferase, go to the full flat file.
Word Map on EC 2.4.1.50 
Reaction
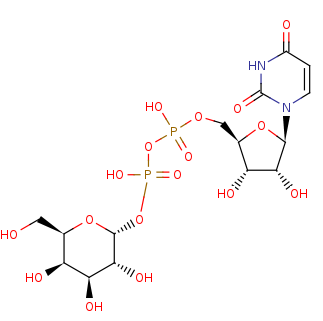 UDP-alpha-D-galactose +
UDP-alpha-D-galactose +
 [procollagen]-(5R)-5-hydroxy-L-lysine=
[procollagen]-(5R)-5-hydroxy-L-lysine=
 UDP +
UDP +
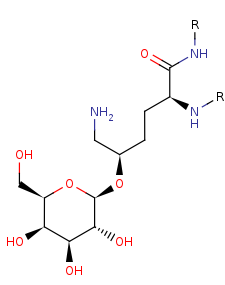 [procollagen]-(5R)-5-O-(beta-D-galactosyl)-5-hydroxy-L-lysine
[procollagen]-(5R)-5-O-(beta-D-galactosyl)-5-hydroxy-L-lysine
Synonyms
B3GALT6, beta(1-O)galactosyltransferase, beta1,O galactosyltransferase GLT25D1, beta3GalT6, COLGALT1, COLGALT2, collagen beta(1-O)galactosyltransferase, collagen galactosyltransferase, collagen galactosyltransferase GLT25D1, collagen hydroxylysyl galactosyltransferase, collagen hydroxylysyl glycosylsyltransferase, galactosyltransferase II, galactosyltransferase, uridine diphosphogalactose-collagen, GLT25D1, GLT25D2, glycosyl transferase25domain 1, hydroxylysine galactosyltransferase, hydroxylysyl galactosyltransferase, LH3, LH3/PLOD3, lysyl hydroxylase 3, multifunctional procollagen lysine hydroxylase and glycosyltransferase, P3H1, PLOD2, PLOD3, procollagen lysyl hydroxylase 2, prolyl 3-hydroxylase 1, UDP galactose-collagen galactosyltransferase, UDPgalactose:5-hydroxylysine-collagen galactosyltransferase, uridine diphosphogalactose-collagen galactosyltransferase
ECTree
Sequence
Sequence on EC 2.4.1.50 - procollagen galactosyltransferase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
PLOD3_HUMAN
738
0
84785
Swiss-Prot
Secretory Pathway (Reliability: 2)
PLOD3_MOUSE
741
0
84922
Swiss-Prot
Secretory Pathway (Reliability: 1)
PLOD3_PONAB
738
0
84785
Swiss-Prot
Secretory Pathway (Reliability: 2)
PLOD3_RAT
741
0
85060
Swiss-Prot
Secretory Pathway (Reliability: 1)
PLOD_CAEEL
730
0
84425
Swiss-Prot
Secretory Pathway (Reliability: 1)
GT251_BOVIN
623
0
71547
Swiss-Prot
Secretory Pathway (Reliability: 1)
GT251_DANRE
604
0
70832
Swiss-Prot
Secretory Pathway (Reliability: 1)
GT251_HUMAN
622
0
71636
Swiss-Prot
Secretory Pathway (Reliability: 2)
GT251_MOUSE
617
0
71061
Swiss-Prot
Secretory Pathway (Reliability: 1)
GT252_HUMAN
626
0
72924
Swiss-Prot
Secretory Pathway (Reliability: 1)
GT252_MOUSE
625
0
72788
Swiss-Prot
Secretory Pathway (Reliability: 2)
G251A_XENLA
611
0
71389
Swiss-Prot
Secretory Pathway (Reliability: 2)
G251B_XENLA
611
0
71607
Swiss-Prot
Secretory Pathway (Reliability: 2)
A0A8B6ED99_MYTGA
245
0
29126
TrEMBL
other Location (Reliability: 5)
K6ZDA6_9ALTE
219
0
25278
TrEMBL
-
A0A6J8EG62_MYTCO
617
0
72433
TrEMBL
other Location (Reliability: 4)
A0A852U3B8_9ACTN
245
0
26864
TrEMBL
-
A0A812DBU1_SEPPH
431
0
50323
TrEMBL
other Location (Reliability: 3)
A0A8M1NI33_DANRE
613
0
70620
TrEMBL
Secretory Pathway (Reliability: 4)
A0A8B6E9Q7_MYTGA
591
0
68654
TrEMBL
Secretory Pathway (Reliability: 1)
A0A6J8BJP4_MYTCO
210
0
24499
TrEMBL
other Location (Reliability: 3)
A0A6J8EJ95_MYTCO
602
0
70719
TrEMBL
other Location (Reliability: 4)
A0A6J8ANJ3_MYTCO
216
0
25463
TrEMBL
other Location (Reliability: 2)
A0A8B6ECK8_MYTGA
225
0
26466
TrEMBL
other Location (Reliability: 1)
A0A8B6DPF0_MYTGA
274
0
32054
TrEMBL
other Location (Reliability: 2)
A0A8B6EC12_MYTGA
606
0
70368
TrEMBL
Secretory Pathway (Reliability: 1)
A0A061IMD6_CRIGR
1099
0
125009
TrEMBL
Mitochondrion (Reliability: 3)
A0A1Z5JMG5_FISSO
278
0
31390
TrEMBL
Secretory Pathway (Reliability: 2)
A0A061IJV9_CRIGR
1088
0
123773
TrEMBL
Mitochondrion (Reliability: 3)
A0A6J7ZW43_MYTCO
259
0
30712
TrEMBL
Secretory Pathway (Reliability: 2)
A0A0C1JUD9_9BACT
308
1
36532
TrEMBL
-
A0A061ING7_CRIGR
1010
0
115607
TrEMBL
Mitochondrion (Reliability: 3)
A0A8B6FUH3_MYTGA
162
0
19073
TrEMBL
other Location (Reliability: 2)
A0A6J8A4U3_MYTCO
283
0
33353
TrEMBL
Secretory Pathway (Reliability: 1)
PLOD2_HUMAN
737
0
84686
Swiss-Prot
-
html completed




 results (
results ( results (
results ( top
top









