1.4.99.6: D-arginine dehydrogenase
This is an abbreviated version!
For detailed information about D-arginine dehydrogenase, go to the full flat file.
Reaction
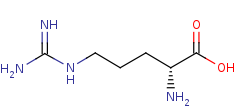 D-arginine +
D-arginine +
 acceptor +
acceptor +
 H2O=
H2O=
 5-guanidino-2-oxopentanoate +
5-guanidino-2-oxopentanoate +
 NH3 +
NH3 +
 reduced acceptor
reduced acceptor
Synonyms
D-alanine dehydrogenase, D-amino acid dehydrogenase, D-amino acid dehydrogenase DadA, DAADH, DAD, DadA, DADH, DauA, EC 1.4.99.1, NADP+-dependent D-amino acid dehydrogenase
ECTree
KM Value
KM Value on EC 1.4.99.6 - D-arginine dehydrogenase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
0.07
2,6-dichlorophenolindophenol
-
specific acceptor, cosubstrate: D-kynurenine
32.3
2-oxo-4-methylvaleric acid
-
at pH 10.5 and 50°C
0.0082
coenzyme Q1
in 50 mM sodium-phosphate buffer (pH 8.0), at 37°C
0.46
D-alanine
pH 8.7, 37°C
0.45
D-kynurenine
-
both methylene blue and 2,6-dichlorophenolindophenol specific enzymes
1.07 - 11
D-phenylalanine
40.2
D-Pro
with Q1 as the electron acceptor, in 50 mM sodium-phosphate buffer (pH 8.0), at 37°C
1.65
D-serine
pH 8.7, 37°C
2.47
D-threonine
pH 8.7, 37°C
0.47
D-tryptophan
-
both methylene blue and 2,6-dichlorophenolindophenol specific enzymes
3.01
D-valine
pH 8.7, 37°C
0.008
methylene blue
-
specific acceptor, cosubstrate: D-kynurenine
0.06
D-arginine

pH 8.7, 25°C
0.06
D-arginine
-
pH 8.7, 25°C
0.06
D-arginine
pH 8.7, 25°C
0.06
D-arginine
mutant Y249F, pH 8.7, 25°C
0.36
D-arginine
mutant Y53F, pH 8.7, 25°C
5.37
D-arginine
pH 8.7, 37°C
0.81
D-histidine

pH 8.7, 37°C
8.8
D-histidine
pH 8.7, 25°C
11
D-histidine
pH 8.7, 25°C
11
D-histidine
-
pH 8.7, 25°C
1.8
D-leucine

wild-type, pH 6.0, 25°C
4.6
D-leucine
wild-type, pH 10.0, 25°C
12
D-leucine
pH 8.7, 25°C
12
D-leucine
-
pH 8.7, 25°C
0.19
D-Lysine

-
-
0.26
D-Lysine
pH 8.7, 25°C
0.26
D-Lysine
-
pH 8.7, 25°C
1.43
D-methionine

-
-
10
D-methionine
pH 8.7, 25°C
10
D-methionine
-
pH 8.7, 25°C
1.07
D-phenylalanine

pH 8.7, 37°C
11
D-phenylalanine
pH 8.7, 25°C
11
D-phenylalanine
-
pH 8.7, 25°C
0.0082
D-proline

Vmax: 12.3 micromol/min/mg
1.82
D-proline
pH 8.7, 37°C
40.2
D-proline
Vmax: 25 micromol/min/mg
0.4
D-tyrosine

-
-
0.8
D-tyrosine
pH 8.7, 25°C
0.8
D-tyrosine
-
pH 8.7, 25°C
1.78
D-tyrosine
pH 8.7, 37°C



 results (
results ( results (
results ( top
top






